NASBA - Nucleic Acid Sequence Based Amplification
The NASBA method
NASBA is a real time enzymatic amplification process that is able to amplify and detect RNA even in the presence of DNA. Unlike PCR, the amplification is run at isothermal conditions at 41°C. Two oligonucleotide primers specific for the RNA target of interest determine the specificity of the reaction. Combined with the use of molecular beacons, that fluoresce only upon hybridisation with their target, real time detection is enabled.
The NASBA reaction continues in a self-sustained manner, thus achieving dramatic amplification in a short period of time.
NASBA components
- Avian Myeloblastosis Virus Reverse Transcriptase (AMV-RT)
- Escherichia coli RNase H
- T7 RNA Polymerase
- Two specific oligonucleotide primers(P1/P2)
- Molecular beacon
- Nucleoside triphosphates,
- Appropriate buffer components
NASBA amplification and detection principles
Primer P1 contains a 5’-terminal T7 RNA polymerase promoter sequence in addition to a stretch of nucleotides that is complementary to a sequence on the target RNA. Primer P2 encompasses a short sequence which is identical to a segment of the target RNA and is located upstream of the region where the first oligonucleotide can anneal.
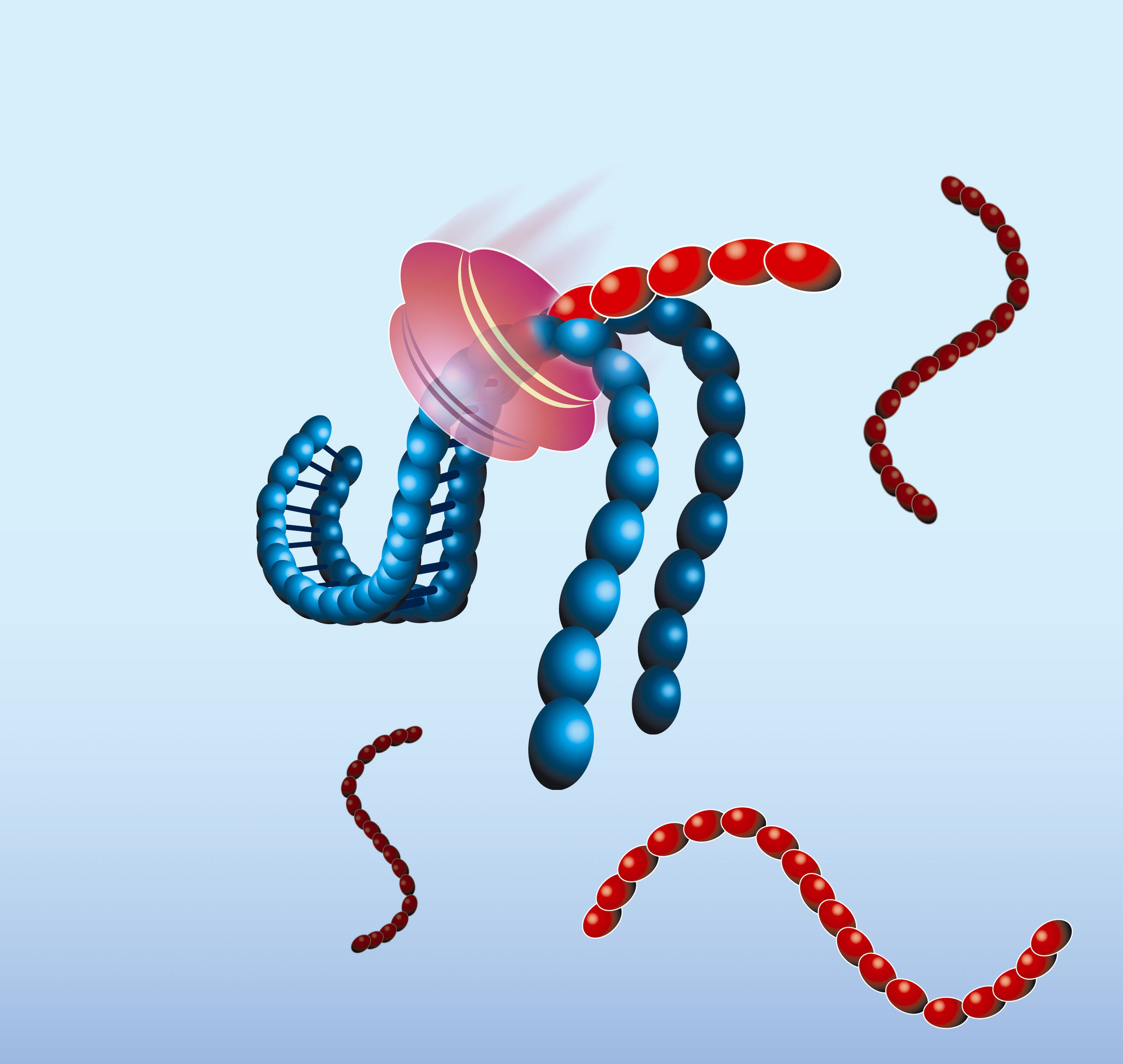
- The NASBA reaction starts with hybridisation of the primer (P1) to the target RNA (red). The primer contains a 5’-terminal T7 RNA polymerase promoter sequence.
- AMV-RT elongates the primer, creating a cDNA copy of the RNA template and forming a RNA/DNA hybrid
- RNase H recognizes this hybrid as substrate and hydrolyses the RNA portion of the hybrid leaving single-stranded DNA. The second primer (P2) anneals to this DNA.
- AMV-RT elongates primer P2 and thus makes the promoter portion of the DNA double stranded and transcriptionally active.
- Recognizing the now functional promoter, T7 RNA Polymerase produces multiple copies of RNA transcripts that are anti-sense to the original target RNA sequence.
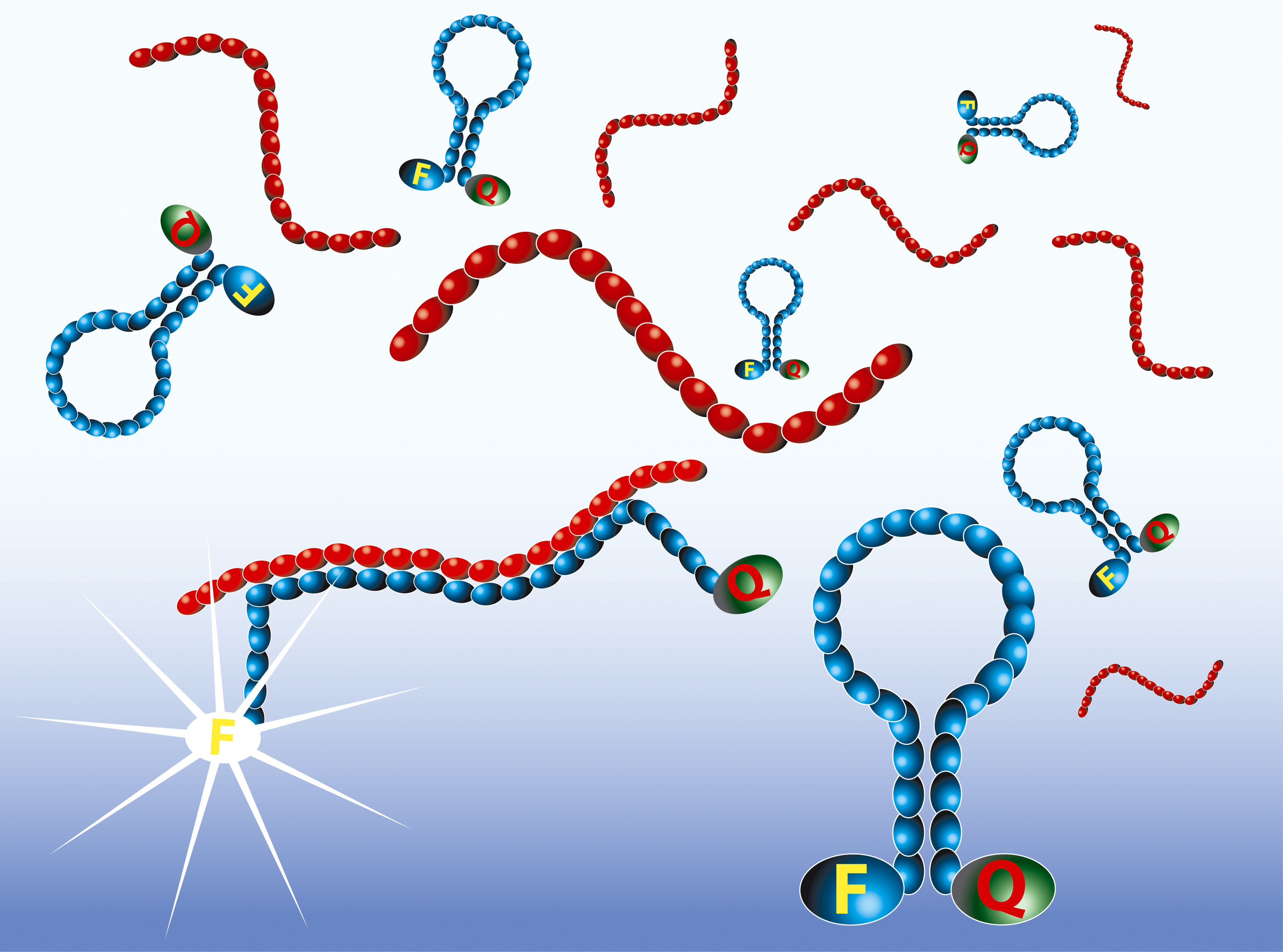
Real-time detection with Molecular Beacons
The amplified RNA is detected in real-time by the use of Molecular Beacon probes. Molecular Beacons are single-stranded oligonucleotides having a stem-loop structure. The loop portion contains the sequence complementary to the target nucleic acid, whereas the stem is unrelated to the target and has a double-stranded structure. One arm of the stem is labelled with a fluorescent dye, and the other arm is labelled with a non-fluorescent quencher. In closed state the probe does not produce fluorescence because the energy is transferred to the quencher. When the loop of the Molecular Beacon hybridises to its target, the Molecular Beacon undergoes a conformational change resulting in a physical separation of the fluorophore and quencher and emission of photons at the wavelength that is specific for the fluorophore.
The intensity of the fluorescence upon hybridisation is related to the amplicon concentration. The real-time measurements show typical exponential curves, with the usual tendency to reach a plateau. The time point at which the fluorescence signal becomes detectable over the background (Time to Positivity) is linear over a range of at least five orders of magnitude of input RNA molecules.